Z03 - Proteomics platform for protein interaction mapping
Principal Investigators
Prof. Dr. Christian Freund, FU Berlin
The focus of Z03 is on supporting the CRC958 with proteomic approaches to delineate interaction partners of membrane scaffolds, to determine post‐translational modifications (PTM) or to profile the intactness and stoichiometry of purified complexes for structural investigations. Typical shotgun proteomic experiments were used in collaboration with projects A03, A13, A19, A21 and A22, with the 18O/16O method becoming a standard procedure to perform quantitative experiments of primary cells or tissue samples. Phosphorylation of Bruchpilot by SRPK79D kinase was investigated in conjunction with A06 (Wahl, Sigrist).
Furthermore, we apply native MS (Figure 1) - where non-denaturing buffers are used in order to preserve the native protein conformation - to investigate protein complexes in their intact form or to analyze complex topologies by tandem MS experiments in which individual protein subunits are released from a larger complex by collision-induced dissociation (CID). In collaboration with A07, native mass spectrometry was utilized to evaluate a possible dimerization of the SH3A domain of intersectin.
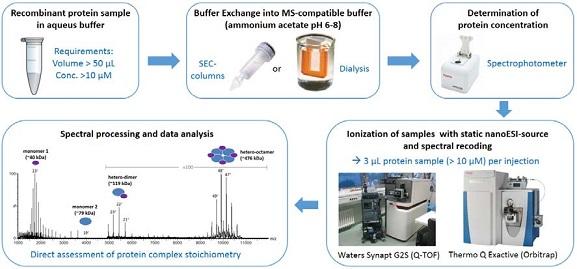
Figure 1: Workflow of Native MS.
Moreover, we use cross-linking MS experiments for interactomic profiling. For instance, by examining neuronal synapses purified from mouse brain or Drosophila heads, we have obtained comprehensive interaction networks in both biological systems. As an illustrative example (Figure 2), we were able to obtain protein interactions together with their binding interfaces of many major proteins/complexes at the synapse. A clear picture emerges from the aggregated dataset: proteins that are known to be localized to synaptic membranes, synaptic vesicles, postsynaptic densities and mitochondria are strongly clustered - a result that is well in agreement with the known spatial organization of these proteins at synapses based on ultra-structural and high-resolution microscopy data.
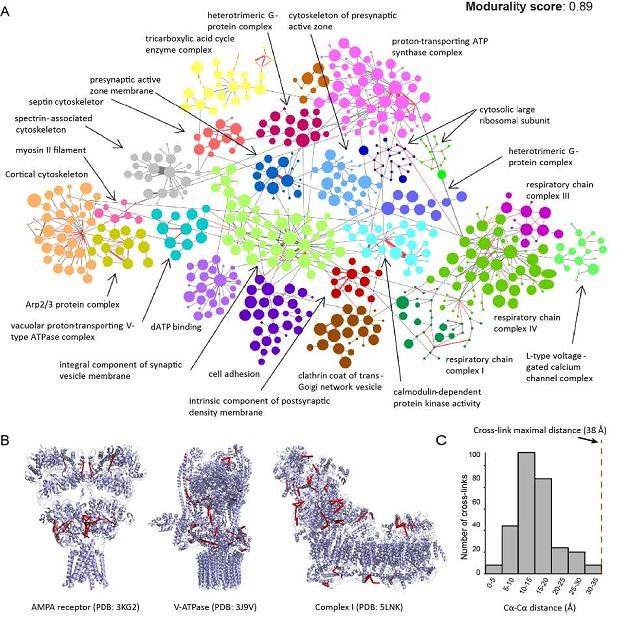
Figure 2: Preliminary results of XL-MS on whole synaptosome preparation. A) network analysis B) structural validation 3) cross-link distance measurement
References:
- Schmerl B, Gimber N, Kuropka B, Stumpf A, Rentsch J, Kunde SA, von Sivers J, Ewers H, Schmitz D, Freund C, Schmoranzer J, Rademacher N, Shoichet SA. The synaptic scaffold protein MPP2 interacts with GABAA receptors at the periphery of the postsynaptic density of glutamatergic synapses. PLoS Biol. 20(3) (2022)
- Jiang PL, Wang C, Diehl A, Viner R, Etienne C, Nandhikonda P, Foster L, Bomgarden RD, Liu F. A Membrane-Permeable and Immobilized Metal Affinity Chromatography (IMAC) Enrichable Cross-Linking Reagent to Advance In Vivo Cross-Linking Mass Spectrometry. Angew Chem Int Ed Engl. e202113937 (2021)
- Lima DB, Zhu Y, Liu F. XlinkCyNET: A Cytoscape Application for Visualization of Protein Interaction Networks Based on Cross-Linking Mass Spectrometry Identifications. J Proteome Res. 20(4):1943-1950 (2021)
- Schnirch L, Nadler-Holly M, Siao SW, Frese CK, Viner R, Liu F. Expanding the Depth and Sensitivity of Cross-Link Identification by Differential Ion Mobility Using High-Field Asymmetric Waveform Ion Mobility Spectrometry. Anal Chem. 92(15):10495-10503 (2020)
- Petzoldt AG, Götz TWB, Driller JH, Lützkendorf J, Reddy-Alla S, Matkovic-Rachid T, Liu S, Knoche E, Mertel S, Ugorets V, Lehmann M, Ramesh N, Beuschel CB, Kuropka B, Freund C, Stelzl U, Loll B, Liu F, Wahl MC, Sigrist SJ. RIM-binding protein couples synaptic vesicle recruitment to release sites. J Cell Biol. 219(7):e201902059 (2020)
- Driller JH, Lützkendorf J, Depner H, Siebert M, Kuropka B, Weise C, Piao C, Petzoldt AG, Lehmann M, Stelzl U, Zahedi R, Sickmann A, Freund C, Sigrist SJ, Wahl MC. Phosphorylation of the Bruchpilot N-terminus in Drosophila unlocks axonal transport of active zone building blocks. J Cell Sci. 132(6):jcs225151 (2019)
- Rademacher N, Kuropka B, Kunde SA, Wahl MC, Freund C, Shoichet SA. Intramolecular domain dynamics regulate synaptic MAGUK protein interactions. Elife. 8:e41299 (2019)
- Gerth F, Jäpel M, Sticht J, Kuropka B, Schmitt XJ, Driller JH, Loll B, Wahl MC, Pagel K, Haucke V, Freund C. Exon Inclusion Modulates Conformational Plasticity and Autoinhibition of the Intersectin 1 SH3A Domain. Structure. 27(6):977-987.e5 (2019)
- Rödel CJ, Otten C, Donat S, Lourenço M, Fischer D, Kuropka B, Paolini A, Freund C, Abdelilah-Seyfried S. Blood Flow Suppresses Vascular Anomalies in a Zebrafish Model of Cerebral Cavernous Malformations. Circ Res. 125(10):e43-e54 (2019)
- Liu F, Lössl P, Rabbitts BM, Balaban RS, Heck AJR. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol Cell Proteomics. 17(2):216-232 (2018)
- Werno MW, Wilhelmi I, Kuropka B, Ebert F, Freund C, Schürmann A. The GTPase ARFRP1 affects lipid droplet protein composition and triglyceride release from intracellular storage of intestinal Caco-2 cells. Biochem Biophys Res Commun. 506(1):259-265 (2018)
- Liu F, Lössl P, Scheltema R, Viner R, Heck AJR. Optimized fragmentation schemes and data analysis strategies for proteome-wide cross-link identification. Nat Commun. 8:15473 (2017)
- Liu Q, Remmelzwaal S, Heck AJR, Akhmanova A, Liu F. Facilitating identification of minimal protein binding domains by cross-linking mass spectrometry. Sci Rep. 7(1):13453 (2017)
- Kuropka B, Schraven B, Kliche S, Krause E, Freund C. Tyrosine-phosphorylation of the scaffold protein ADAP and its role in T cell signaling. Expert Rev Proteomics. (6):545-54 (2016)
- Morrison E, Kuropka B, Kliche S, Brügger B, Krause E, Freund C. Quantitative analysis of the human T cell palmitome. Sci Rep. 5:11598 (2015)
-
Barucker C, Sommer A, Beckmann G, Eravci M, Harmeier A, Schipke CG, Brockschnieder D, Dyrks T, Althoff V, Fraser PE, Hazrati LN, George-Hyslop PS, Breitner JC, Peters O, Multhaup G. Alzheimer amyloid peptide aβ42 regulates gene expression of transcription and growth factors. J. Alzheimers Dis. 44, 613-24 (2015)
-
Kuropka, B., Royla, N., Freund, C., Krause, E. Sortase A mediated site-specific immobilization for identification of protein interactions in affinity purification-mass spectrometry experiments. Proteomics. 15, 1230-1234 (2015)
- Liu F, Rijkers DT, Post H, Heck AJ. Proteome-wide profiling of protein assemblies by cross-linking mass spectrometry. Nat Methods.(12):1179-84 (2015)
- Baumeier C, Kaiser D, Heeren J, Scheja L, John C, Weise C, Eravci M, Lagerpusch M, Schulze G, Joost HG, Schwenk RW, Schürmann A. Caloric restriction and intermittent fasting alter hepatic lipid droplet proteome and diacylglycerol species and prevent diabetes in NZO mice. Biochim Biophys Acta. 1851(5):566-76 (2015)
-
Eravci, M., Sommer, C.& Selbach, M. IPG strip-based peptide fractionation for shotgun proteomics. Methods Mol. Biol. 1156, 67-77 (2014)
- Stephanowitz, H., Lange, S., Lang, D., Freund, C. & Krause, E. An improved two-dimensional reversed phase-reversed phase LC-MS/MS approach for identification of peptide-protein interactions. J. Proteome Res. 11, 1175-83 (2012)
- Lange, S., Sylvester, M., Schümann, M., Freund, C. & Krause, E. Identification of phosphorylation-dependent interaction partners of the adapter protein ADAP using quantitative mass spectrometry: SILAC vs (18)O-labeling. J. Proteome Res. 9, 4113-22 (2010)
-
Lehmann, R., Meyer, J., Schuemann, M., Krause, E. & Freund, C. A novel S3S-TAP-tag for the isolation of T-cell interaction partners of adhesion and degranulation promoting adaptor protein. Proteomics 9, 5288-5295 (2009)
-
Kofler, M., Schümann, M., Merz, C., Kosslick, D., Schlundt, A., Tannert, A. Schaefer, M., Lührmann, R., Krause, E. and Freund, C. Proline-rich sequence recognition I: Marking GYF and WW domain assembly sites in early spliceosomal complexes. Mol. Cell. Proteom. 8 2461-2473 (2009)
-
Schlundt A, Sticht J, Piotukh K, Kosslick D, Jahnke N, Keller S, Schuemann M, Krause E, Freund C. Proline-rich sequence recognition II: proteomic analysis of TSG101-UEV interactions. Mol. Cell. Proteom. 8, 2474-2486 (2009)