A04 - Spatiotemporal model of neuronal signalling and its regulation by presynaptic membrane scaffolds
Principal Investigator
Prof. Dr. Frank Noé, FU Berlin
The specific arrangement of proteins, lipids and other molecules in space and time is a prerequisite for the precise and efficient signaling at membrane scaffolds. In this project, we develop theoretical and computational models that describe the detailed spatiotemporal arrangement of proteins, membranes, and their quantitative orchestration in presynaptic neuronal exo- and endocytosis.
We have developed methods that facilitate data integration and simulation of spatiotemporal models of membrane scaffolds. Our workhorse is the reaction-diffusion software ReaDDy. We have also developed methods to estimate the slowly-interconverting conformations of macromolecules from molecular dynamics simulations or single-molecule data. Finally, we have contributed to the improvement of image recovery from STORM microscopy in collaboration with the Haucke/Maritzen (A01) and Schmoranzer (Z02) labs.
We have applied our methods in order to study selected spatiotemporal effects of exo/ endocytic signaling in neurons. On the exocytic side, we have found that the dynamical cluster formation of the SNARE protein syntaxin can be explained with a simple physical model. In collaboration with the Sigrist lab (A03), we have found that syntaxin clusters tend to grow larger and preferentially locate at active zones, thus likely facilitating efficient docking of vesicles. On the endocytic side, we have modeled the phospholipid-dependent recruitment of proteins such as SNX9 to clathrin-coated pits and studied the spatiotemporal arrangement of membrane remodeling proteins, in particular SNX9 and dynamin. These studies are in collaboration with the labs of Haucke/Maritzen, Schmoranzer/Ewers and Daumke (A01, Z02, A12).
In the current funding period, we continue to build up a spatiotemporal model of exo- endocytosis. In particular, we are extending our reaction-diffusion model by developing a flexible membrane model that captures balance of binding/dissociation of membrane-remodeling proteins such as SNX9 and dynamin and also the associated membrane curvature changes. These works are in close collaboration with Haucke/Maritzen and Schmoranzer/Ewers (A01, Z02).
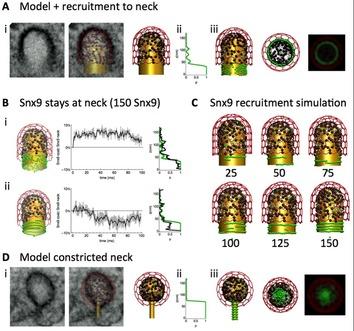
Figure 1: Spatiotemporal ReaDDy model and simulations of dome-shaped (A-C) and constricted (D) clathrin coated pits (CCPs). Color-coding: Membrane (gold), SNX9 (green), clathrin (red), other CCP proteins (black). EM micrographs (Ai, Di) were used to model the CCP membrane before and after constriction.
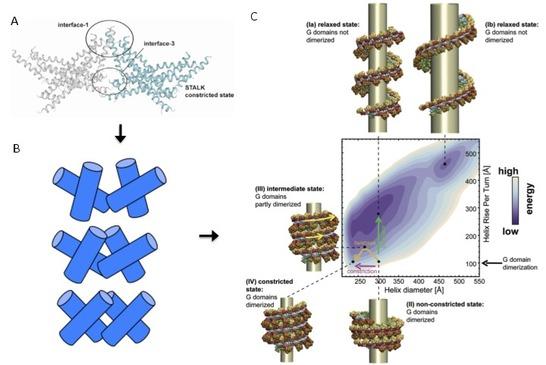
Figure 2: Atomistic MD simulation setup of the stalk tetramer (180.000 atoms with solvent). B: Schematic of set of stalk tetramers sampled in the simulation. C: Computational free energy model of the dynamin oligomer successfully predicting the experimentally known relaxed and constricted states, and suggesting a sequence of constriction and relaxation leading to membrane rupture.
References:
-
Sadeghi, M., Weikl, T. R. and F. Noé. Particle-based membrane model for mesoscopic simulation of cellular dynamics. J. Chem. Phys.148: 044901 (2018)
-
Mardt, A., Pasquali, L., Wu, H. and F. Noé. VAMPNets for deep learning of molecular kinetics. Nat. Commun. 9: 5 (2018)
-
Fröhner C. and F. Noé. Reversible interacting-particle reaction dynamics. J. Phys. Chem B Article ASAP, J. Phys. Chem. B, Article ASAP, DOI: 10.1021/acs.jpcb.8b06981 (2018)
-
Paul, F., Wehmeyer, C., Abualrous, E.T., Wu, H., Crabtree, M. D., Schöneberg, J., Clarke, J., Freund, C., Weikl, T. R. and F. Noé. Protein-peptide association kinetics beyond the seconds timescale from atomistic simulations. Nat. Commun. 8: 1095 (2017)
-
Plattner N., Doerr S., De Fabritiis G. and F. Noé. Complete protein–protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem. 9: 1005–1011 (2017)
-
Schöneberg, J., Lehmann, M., Ullrich, A., Posor, Y., Lo, W. T., Lichtner, G., Schmoranzer, J., Haucke, V. and F. Noé. Lipid-mediated PX-BAR domain recruitment couples local membrane constriction to endocytic vesicle fission. Nat. Commun. 8: 15873 (2017)
-
Albrecht, D., Winterflood, C. M., Sadeghi, M., Tschager, T., Noé, F., and H. Ewers. Nanoscopic compartmentalization of membrane protein motion at the axon initial segment. J. Cell. Biol. 215: 37-46 (2016)
-
Ullrich, A., Böhme, M. A., Schöneberg, J., Depner, H., Sigrist, S. J. and Noé, F. Dynamical organization of Syntaxin-1A at the presynaptic active zone. PLoS Comput. Biol. 11. e1004407 (2015)
-
Reubold, T. F., Faelber, K., Plattner, N., Posor, Y., Branz, K., Curth, U., Schlegel, J., Anand, R., Manstein, D., Noé, F., Haucke, V., Daumke, O. and Eschenburg, S. Crystal structure of the dynamin tetramer. Nature 525. 404-408 (2015)
-
Biedermann, J., Ullrich, A., Schöneberg, J. and Noé, F. ReaDDyMM: fast interacting-particle reaction-diffusion simulations using graphical processing units. Biophys. J. 108. 457-461 (2015)
-
Schöneberg, J. and Noé, F. ReaDDy - a software for particle based reaction diffusion dynamics in crowded cellular environments. PLoS ONE 8, e74261 (2013)
-
Posor, Y., Eichhorn-Gruenig, M., Puchkov, D., Schöneberg, J., Ullrich, A., Lampe, A., Müller, R., Zarbakhsh, S., Gulluni, F., Hirsch, E., Krauss, M., Schultz, M. C., Schmoranzer, J., Noé, F. and Haucke, V. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature 499, 233-237 (2013)
-
Faelber, K., Held, M., Gao, S., Posor, Y., Haucke, V., Noé, F. and Daumke, O. Structural insights into dynamin-mediated membrane fission. Structure 20, 1621-1628 (2012)
-
Faelber, K., Posor, Y., Held, M., Roske, Y., Schulze, D., Haucke, V., Noé, F. and Daumke, O. Crystal structure of nucleotide-free dynamin. Nature 477, 556-560 (2011)