Z05 - Three-dimensional cryo-electron microscopy
Prinzipal Investigator
Prof. Dr. Christian Spahn, Charité Berlin
Recent revolutionary technological and methodological advances made three‐dimensional cryoelectron microscopy (3D cryo‐EM) a key‐method for the structural analysis of macromolecular
assemblies and machines and for visualizing the architectures of sub‐cellular compartments at unprecedented resolution. Single‐particle cryo‐EM analyses (SPA cryo‐EM) now make it possible to
determine the structure of isolated macromolecular assemblies at near‐atomic resolution. In combination with multiparticle, in silico sorting approaches it allows the analysis even of conformationally or compositionally heterogeneous samples and thus can provide insight into intrinsicstructural variability or dynamic conformational modes.
While SPA cryo‐EM relies on averaging of particle images, cryo‐electron tomography (cryo‐ET) (Figure 1) allows the imaging of unique objects such as organelles or entire cells. In combination with micromachining using a dual beam instrument with focused ion beam and scanning electron microscope (cryo‐FIBSEM) cryo‐ET facilitates visualizing subcellular compartments and macromolecular assemblies in the native environment of the cell at nanometer to sub‐nanometer resolution.
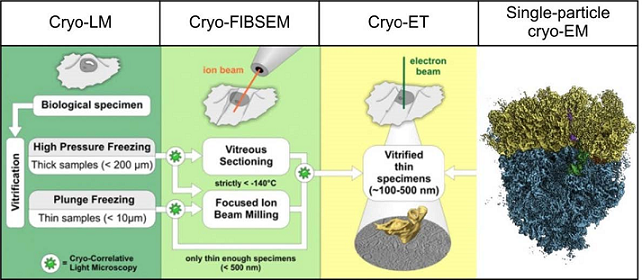
Figure 1: Overview of 3D cryo-EM methodology. The left three columns show the cryo-electron tomography (cryo-ET) workflow encompassing cryo-correlative light microscopy (cryo-LM), thinning of the specimen using a dual beam cryo-FIBSEM instrument combining a focused ion beam (FIB) and a scanning electron microscope (SEM) and reconstruction of 3D volume from a recorded tilt-series by cryo-ET (adopted with modifications from ref 4). The right column exemplifies SPA cryo-EM at near-atomic resolution by the first structure of the human 80S ribosome (Behrmann et al., 2015). SPA reconstructions can be further used to identify macromolecular complexes in cryo-ET reconstructions for interpretation and further image processing steps such as subtomogram averaging.
The overall aim of project Z5 is to facilitate 3D cryo‐EM analyses within the SFB958 consortium. In close collaboration, we will provide SPA cryo‐EM or molecular cryo‐ET to visualize macromolecular assemblies provided by the individual projects of the consortium. For example, we want to continue our collaboration with project A16 (Eickholt) do determine the SPA cryo‐EM structure of PRG2 oligomers and we want to build on the recent successful application for high‐end cryo‐EM instruments (high-end 300 kV cryo‐TEM, cryo‐FIBSEM, cryo‐LM) and implement the workflow for cellular cryo‐ET to study the structure of Drosophila synaptosomes with project A03 (Sigrist). Furthermore, we want to use molecular cryo‐ET to study protein‐induced membrane tubule formation with projects A07 (Freund, Haucke) and A12 (Daumke).
For instance, in close collaboration with A12 (Daumke), we analyse structural aspects of Eps15 homology domain containing proteins (EHDs), a family of dynamin-related ATPases involved in membrane trafficking. ATP-dependent oligomerization of EHDs can remodel membranes resulting in EHD-decorated tubules18 (Shah et al., 2014). Fourier-Bessel helical reconstruction methods or the SPA inspired Iterative Helical Real Space Reconstruction (IHRSR)19, 20 approach may be suited in principle to determine the structure of such tubules. However, the strong heterogeneity, e.g. varying diameters, of membrane tubules, decorated with EHD2 or the related EHD4ΔN, respectively, renders the application of these approaches prohibitively difficult. A couple of years ago, we therefore explored the possibility to use molecular cryo- ET in combination with subtomogram veraging. In a pilot experiment, tomographic tilt series of EHD4ΔN covered membrane tubules were collected at a Krios Titan 300 kV Cryo-TEM the EMBL in Heidelberg. The initial tomographic reconstruction revealed the striking plasticity of the tubules. A preliminary subtomogram average revealed regular rows of density that, based on the X-ray structure of EHD4ΔN21, can be identified as oligomeric EHD4 (Figure 2). These results were highly encouraging and paved the way for further in-depth analysis of EHD-induced membrane tubule formation and other oligomerization processes of scaffold proteins at membranes and our paper "Cryo-electron tomography reveals structural insights into the membrane binding and remodeling activity of dynamin-like EHDs." will be published soon.
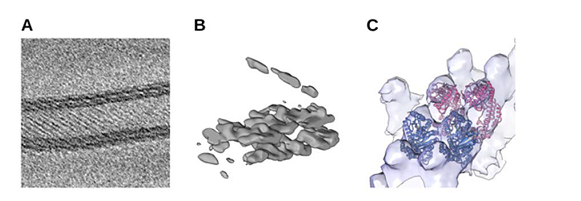
Figure 2: Preliminary subtomogram average of EHD4ΔN. (A) Slice through a tomographic reconstruction of oligomeric EHD4ΔN on a membrane, showing typical striations. (B) A single subtomogram identified with Dynamo22 after strong low-pass filtering shows a section through the membrane. (C ) Cryo-EM density after subtomogram averaging superposed with atomic models of EHD4ΔN•ATP-γS in the active state21 (PDB 5MTV).
References:
- Kraushar ML, Krupp F, Harnett D, Turko P, Ambrozkiewicz MC, Sprink T, Imami K, Günnigmann M, Zinnall U, Vieira-Vieira CH, Schaub T, Münster-Wandowski A, Bürger J, Borisova E, Yamamoto H, Rasin MR, Ohler U, Beule D, Mielke T, Tarabykin V, Landthaler M, Kramer G, Vida I, Selbach M, Spahn CMT. Protein Synthesis in the Developing Neocortex at Near-Atomic Resolution Reveals Ebp1-Mediated Neuronal Proteostasis at the 60S Tunnel Exit. Mol Cell. 81(2):304-322.e16 (2021)
- Grunwald S, Hopf LVM, Bock-Bierbaum T, Lally CCM, Spahn CMT, Daumke O. Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates. Nat Commun. 11(1):5506 (2020)
- Brosig A, Fuchs J, Ipek F, Kroon C, Schrötter S, Vadhvani M, Polyzou A, Ledderose J, van Diepen M, Holzhütter HG, Trimbuch T, Gimber N, Schmoranzer J, Lieberam I, Rosenmund C, Spahn C, Scheerer P, Szczepek M, Leondaritis G, Eickholt BJ. The Axonal Membrane Protein PRG2 Inhibits PTEN and Directs Growth to Branches. Cell Rep 29(7):2028-2040.e8 (2019)
- Qureshi, B.M., A. Schmidt, E. Behrmann, J. Bürger, T. Mielke, C.M.T. Spahn, M. Heck, and P. Scheerer. Mechanistic insights into the role of prenyl-binding protein PrBP/δ in membrane dissociation of phosphodiesterase 6. Nat Commun. 9:90 (2018a)
- Qureshi BM, Behrmann E, Schöneberg J, Loerke J, Bürger J, Mielke T, Giesebrecht J, Noé F, Lamb TD, Hofmann KP, Spahn CMT, Heck M. It takes two transducins to activate the cGMP-phosphodiesterase 6 in retinal rods. Open Biol. 8(8):180075 (2018b)
- Nikolay R, Hilal T, Qin B, Mielke T, Bürger J, Loerke J, Textoris-Taube K, Nierhaus KH, Spahn CMT. Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution. Mol Cell. 70(5):881-893.e3 (2018)
- Said, N., F. Krupp, E. Anedchenko, K.F. Santos, O. Dybkov, Y.H. Huang, C.T. Lee, B. Loll, E. Behrmann, J. Bürger, T. Mielke, J. Loerke, H. Urlaub, C.M.T. Spahn, G. Weber, and M.C. Wahl. Structural basis for λN-dependent processive transcription antitermination. Nat Microbiol. 28;2:17062 (2017)
- Hilal, T., H. Yamamoto, J. Loerke, J. Bürger, T. Mielke, and C.M. Spahn. Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution. Nat Commun. 20;7:13521 (2016)
- Sprink, T., D.J. Ramrath, H. Yamamoto, K. Yamamoto, J. Loerke, J. Ismer, P.W. Hildebrand, P. Scheerer, J. Bürger, T. Mielke, and C.M. Spahn. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Science Adv. 2(3): e1501502 (2016)
- Behrmann, E., J. Loerke, T.V. Budkevich, K. Yamamoto, A. Schmidt, P.A. Penczek, M.R. Vos, J. Bürger, T. Mielke, P. Scheerer, and C.M.T. Spahn. Structural snapshots of actively translating human ribosomes. Cell. 161: 845 – 857 (2015)
- Shah, C., B.G. Hegde, B. Morén, E. Behrmann, T. Mielke, G. Moenke, C.M.T. Spahn, R. Lundmark, O. Daumke, and R. Langen. Structural insights into membrane interaction and caveolar targeting of dynamin-like EHD2. Structure. 22: 409-20 (2014)
- Fritsch, J., P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz, and C.M.T. Spahn. The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature. 479: 249-52 (2011)
- Penczek, P.A., M. Kimmel, and C.M.T. Spahn. Identifying conformational states of macromolecules by eigen-analysis of resampled cryo-EM images. Structure. 19: 1528-90 (2011)
Model of splicing-regulated secretion modulators. Alternative splicing controls all steps of the secretory pathway. Selected candidates and their localization in the cell are shown. Alternative splicing of candidates in red has been confirmed to control proteins secretion using the RUSH system.